Understanding why we sleep remains one of the most enduring mysteries in science. A core feature of sleep is its homeostatic regulation, i.e., waking experience drives subsequent sleep. Here we are investigating the molecular basis of the sleep homeostat: 1) how does the homeostat sense waking signals? 2) How does the homeostat trigger sleep? 3) How does the circadian clock interact with the sleep homeostat? 4) How does sleep promote optimal brain function and protect against disease. We approach these questions using a variety of -omics approaches with a corresponding need for analytical methods.
Nearly every organism examined, even the jellyfish that lacks a centralized nervous system exhibits a restorative sleep-like state. While asleep, we cannot eat, mate, defend ourselves from predators or care for our young. Inadequate sleep contributes to brain disease such as Alzheimer’s and depression and even diseases outside of the brain, such as diabetes and obesity. Sleep is homeostatically regulated, i.e., sleep is driven by the duration and intensity of prior waking experience. Our principal goals are to identify the neuronal and molecular components of the sleep homeostat, to understand how those components cooperate to sense and control sleep-wake state, and to reveal how molecular and neural homeostatic pathways impact brain function, health, and disease. Our strategy employs genetic tools primarily in the fruit fly Drosophila to dissect, manipulate, and monitor the homeostatic machinery. This approach builds on our research to understand the molecular basis of circadian (~24 h) behavior which have revealed sleep mechanisms conserved between invertebrates and vertebrates and work in both mice and humans.
Circadian Rhythms
Circadian (~24 h) clocks dictate when we wake up and when we fall asleep. Using molecular to behavioral approaches primarily in the fruit fly, we aim to reveal the molecular and neuronal mechanisms by which circadian clocks keep time and convey that information to control sleep/wake? We are also determining the mechanistic role of disrupted circadian clocks in neurodevelopmental (e.g. autism) and neurodegenerative (e.g. Huntington’s disease) disorders.
Sleep Homeostasis
The elusive sleep homeostat drives sleep as a function of prior wakefulness. How does the homeostat sense waking experience, trigger sleep and restore the brain to a baseline healthy state? By combining genetics, genomics, real-time imaging, and high-resolution behavior analysis, we aim to identify the locus of the sleep homeostat and understand the molecular mechanisms that govern homeostat function. We are also determining how disrupted sleep can contribute to neurodegenerative disorders such as Alzheimer’s disease.
Humans: Diagnostics/Jet Lag
Disrupted sleep and circadian rhythms have been increasingly associated with neural (depression and Alzheimer’s) and even non-neural disorders (diabetes and obesity). Here we are combining RNA-sequencing with machine learning algorithms to discover biomarker signatures of sleep and circadian disorders. We are also using publicly available data to assess the effects of sleep/circadian disruption on athletic performance.
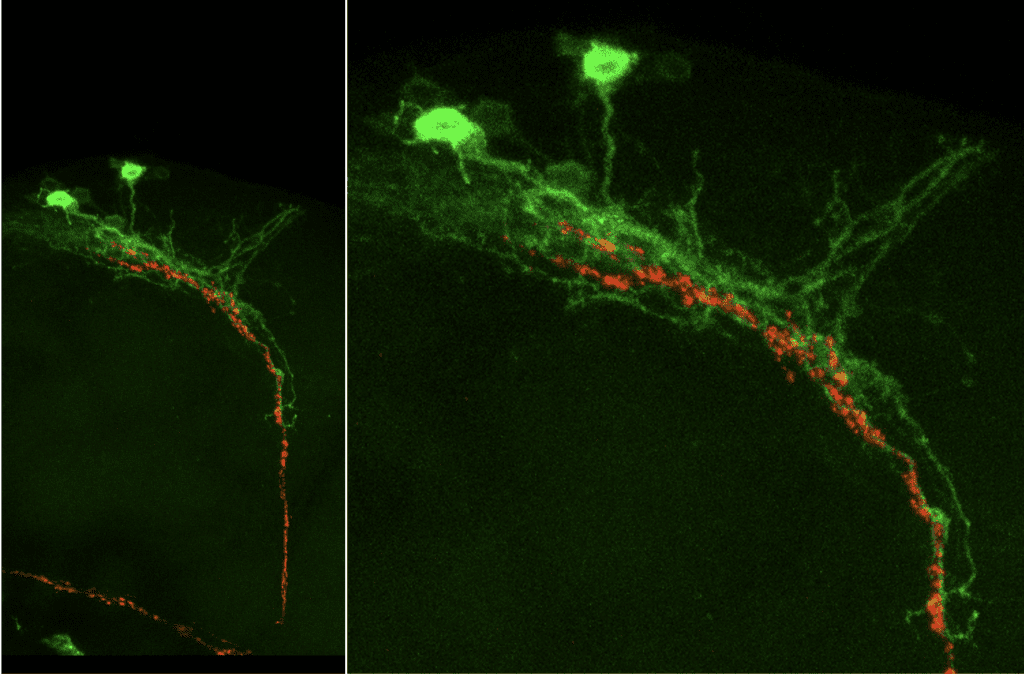
Synaptic connections between circadian clock neurons detected by GFP reconstitution across synaptic partners (GRASP)
Two of my most interesting projects :
1. Discovery of novel human blood biomarkers for circadian rhythms and sleep
Disrupted sleep and circadian rhythms have been increasingly associated with neural (depression and Alzheimer’s) and even non-neural disorders (diabetes and obesity). Here we are combining RNA-sequencing with machine learning algorithms to discover biomarker signatures of sleep and circadian disorders. We collect blood samples under controlled lab conditions or in the field and subject these samples RNA sequencing to quantify gene expression across the entire genome (transcriptome) and metabolomics to quantify metabolites. We then apply machine learning techniques, such as elastic net regression, to “predict” internal circadian time from gene expression. We are currently trying to broaden these techniques to assess other circadian features, such as amplitude, synchrony or strength, and their relationship to organ-specific functions, as well as to sleep drive.
2. Discovery of evolutionarily conserved signatures of sleep across diverse phyla
Understanding why we sleep remains one of the most enduring mysteries in science. A core feature of sleep is its homeostatic regulation, i.e., waking experience drives subsequent sleep. Yet the underlying molecular basis of the sleep homeostat remains unclear. Nearly every organism examined, even the jellyfish that lacks a centralized nervous system exhibits a restorative sleep-like state. We hypothesize that sleep evolved hundreds of millions of years ago and thus the core features of the sleep homeostat are widely conserved. To address this question, we will assemble, from published datasets and those that our lab has generated, large-scale -omics studies that have examined sleep wake datasets. We will then use a variety of analytical approaches as well as to extract those genetic pathways that exhibit sleep-wake dependence invertebrate and vertebrate species to discover these universal signatures.
On my way to becoming a medical doctor, I decided to take a year (and then two) off during medical school to do research at the NIH. There, I caught the research “bug” and began my career-long interest in sleep, working on mechanisms of general anesthesia in the fruit fly Drosophila. After completing my M.D. and a short residency in Clinical Pathology, I did my postdoctoral training with Nobel laureate Michael Rosbash, cloning the core circadian clock gene Clock in Drosophila. I joined the faculty at Northwestern in 2000 where my lab has discovered core gears of the circadian clock, how those gears drive sleep and wake, and how those pathways are linked to neurodevelopmental, neuropsychiatric, and neurodegenerative diseases. I have also been applying similar approaches to reveal the molecular basis of the sleep homeostat, key to understanding the elusive function of sleep. In 2023, I joined the University of Michigan as the Executive Director of the Michigan Neuroscience Institute.
I would like to discover novel biomarker signatures that will both enable a fundamental understanding of sleep, diagnoses of sleep and circadian disorders, and ultimately novel therapeutics.
I am excited by the extraordinary potential of data science and AI research in mining large datasets to discover the needles in the haystack.
I play basketball with some of the other faculty and students and leverage my interest in athletics, examining the impact of jet lag on athletic performance initially using Major League Baseball datasets.
