Larson’s research has been in the area of “Complex Fluids,” which include polymers, colloids, surfactant-containing fluids, liquid crystals, and biological macromolecules such as DNA, proteins, and lipid membranes. He has also contributed extensively to fluid mechanics, including microfluidics, and transport modeling. He has also has carried out research over the past 16 years in the area of molecular simulations for biomedical applications. The work has involved determining the structure and dynamics of lipid membranes, trans-membrane peptides, anti-microbial peptides, the conformation and functioning of ion channels, interactions of excipients with drugs for drug delivery, interactions of peptides with proteins including MHC molecules, resulting in more than 50 publications in these areas, and in the training of several Ph.D. students and postdocs. Many of these studies involve heavy use of computer simulations and methods of statistical analysis of simulations, including umbrella sampling, forward flux sampling, and metadynamics, which involve statistical weighting of results. He also has been engaged in analysis of percolation processes on lattices, including application to disease propagation.
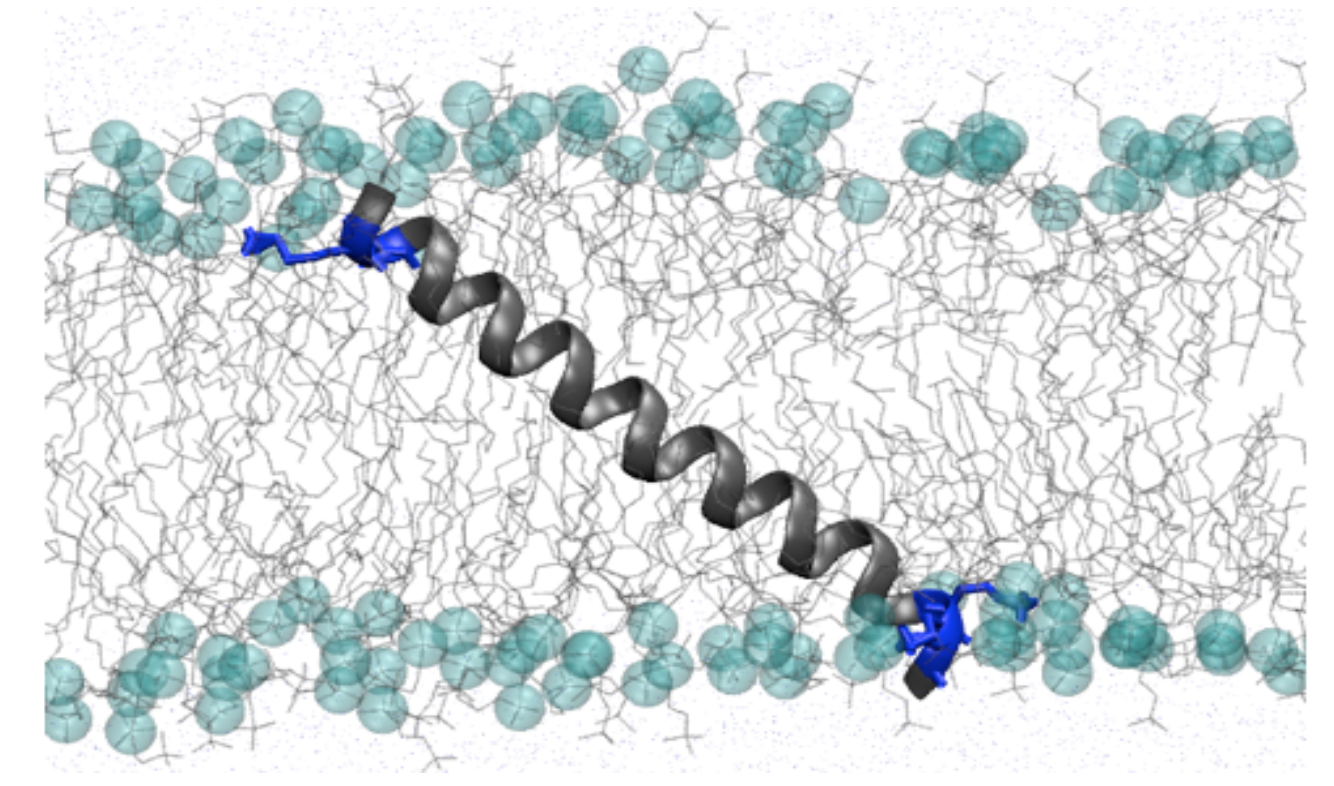
Alpha helical peptide bridging lipid bilayer in molecular dynamics simulations of “hydrophobic mismatch.”
